Case Studies
How about going next with us?
For more information, please feel free to contact us from our inquiry
form!!

WHY NOT EXPLORE THE FUTURE TOGETHER?by HPC SYSTEMS

WHY NOT EXPLORE THE FUTURE TOGETHER?by HPC SYSTEMS

Upcoming exhibition at 2025 MRS Spring (Seattle, April, Booth No. 211)

Releaseed "GRRM®map"!!

Refining Chemical Reaction Mechanisms with Quantum Chemistry for Reaction-Flow Analysis

Establishing Highly Precise Quantitative Analysis Method for Valuable Compound Utilizing Quantum Chemical Calculation

Benchmark of the 5th Generation Intel Xeon Scalable Processor (Emerald Rapids)

Promotion at GDCh Conference on Inorganic Chemistry (Germany, September)
GRRM®23
Automatic chemistry program for exploring all reaction paths and molecules
Reaction plus
Pro 2 / Express 2
Original calculation software for unraveling mechanisms of target chemical reactions

Gaussian
De facto standard software of quantum chemistry calculations

Science Cloud
Cloud platform for starting your calculations readily in HPC environment with pre-installed science software





GRRM®23
Automatic chemistry program for exploring all reaction paths and molecules

Reaction plus
Pro 2 / Express 2
Original calculation software for unraveling mechanisms of target chemical reactions

Gaussian
De facto standard software of quantum chemistry calculations

Science Cloud
Cloud platform for starting your calculations readily in HPC environment with pre-installed science software

Upcoming exhibition at 2025 MRS Spring (Seattle, April, Booth No. 211)

Releaseed "GRRM®map"!!

Refining Chemical Reaction Mechanisms with Quantum Chemistry for Reaction-Flow Analysis

Establishing Highly Precise Quantitative Analysis Method for Valuable Compound Utilizing Quantum Chemical Calculation

Benchmark of the 5th Generation Intel Xeon Scalable Processor (Emerald Rapids)

Promotion at GDCh Conference on Inorganic Chemistry (Germany, September)
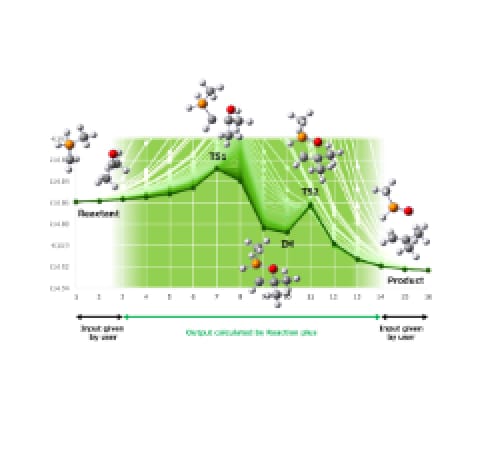
Chemical Reaction Software
Mechanism finder of chemical reactions
Reaction plus Pro 2/Express 2 is our original software that supports you to
identify chemical reaction pathways automatically just by inputting information of reactants and
products. This program was developed to solve the extreme difficulty of optimizing transition state
structures during reaction pathways which fully requires experiences and intuitions of experts in the
conventional method.
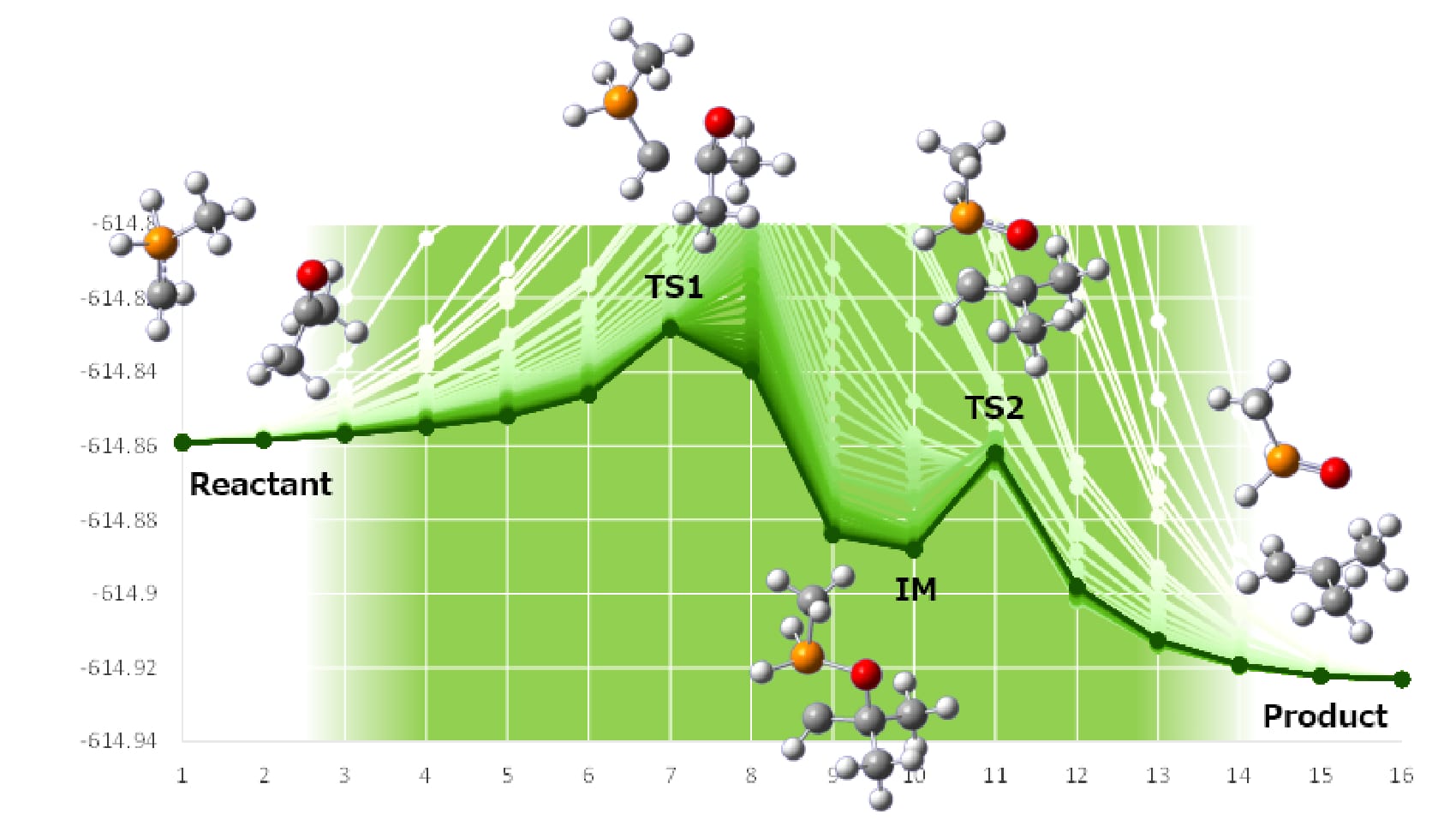
In the latest version of Reaction plus, Pro2/Express2 (meaning version 2),
calculations of various reaction systems are additionally able to be performed from the previous
version, such as enzyme reactions by the ONIOM method, heterogenous catalytic reactions, and open-shell
reactions.
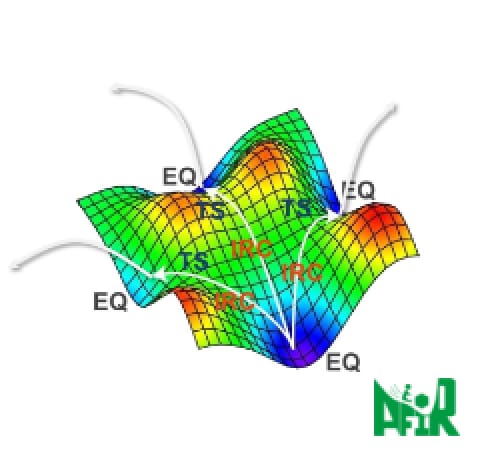
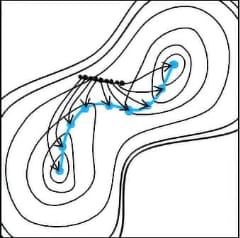
In the conventional optimization method of transition states, a structure close to that of the target transition state has to be specified as the initial structure, which requires a craftsmanship level of techniques. This method causes exhaustive cases that the obtained TS had nothing to do with the reaction after vibration analyses, or that IRC calculations did not run well from TS and eventually the reaction pathways could not be obtained.
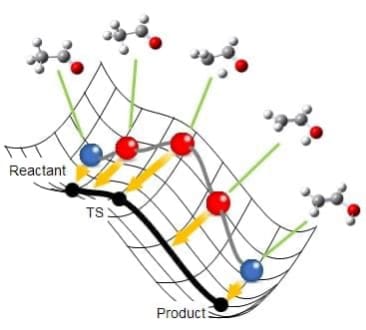
Reaction plus optimizes the entire reaction path based on the Nudged Elastic Band (NEB) method.
This enables us to obtain the transition state structure on the reaction pathway without any
difficulty.
Reaction plus uses the NEB method and String method to optimize the entire rection
path.
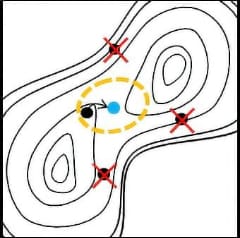
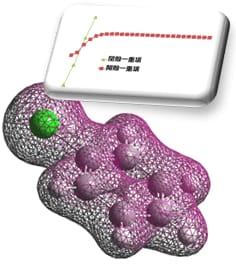
If we compare the molecular structure to a single bead, the reaction path can be expressed
as a string of beads. When this string rolls on the reaction potential curved surface, the reaction
path of the string finally becomes a solution due to the balance between the force that each bead
tries to roll in the direction of decreasing energy and the binding force of the string. Converge to
this is the basic mechanism for reaction path optimization.
The basic mechanism of Reaction plus
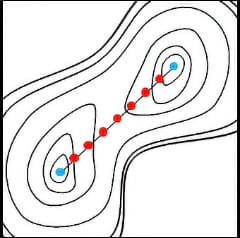
At first, initial and final states of chemical structures are specified by the users, and beads (chemical structures) will be placed at equal intervals between the two inputs, forming a string of beads.
The shape of the string (positions of the beads) will be optimized according to movements of each bead driven by the forces, which means to be a conversion to the target reaction pathway. At this time, the beads avoid approaching too close together by moving toward orthogonal direction toward the string.
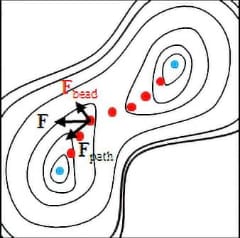
The optimization will be finished when the beads stop their movement. Since the direction of the force against each bead are limited to the direction along the string (the reaction pathway), this implies the pathway is successfully solved.
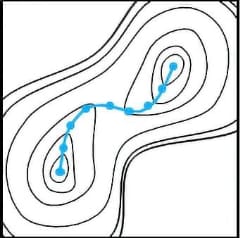
The molecule structures of initial and final state don’t have to be stable structures necessarily. Therefore, users are also able to solve the reaction pathways by specifying the structures instinctively from their past experiences and intuitions.
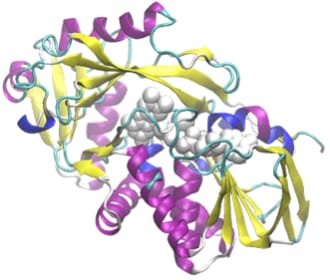
[Adapted to ONIOM method]
ONIOM method is suitable to ensure both calculation accuracy and
speed, which computes minutely to reaction active site but roughly to surroundings. By applying this
method, calculations of large-scale molecules such as enzyme reactions are now available.
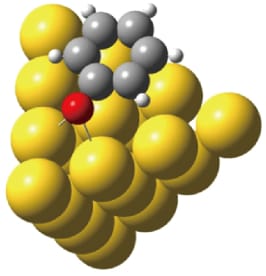
[Atomic coordinate fixing function for homogeneous reactions]
Reaction simulation can be
performed by fixing coordinates of particular atom. This function enables calculations of
homogeneous reaction systems such as catalytic reactions on metal surface.
[Reading function of molecular orbit adapting to open-shell reactions]
By using Gaussian
checkpoint (chk) file, designations of initial orbit to molecular structures on reaction pathways
can be done (initial guess). This allows you to handle reactions of complex open-shell systems such
as radical dissociations and intersystem crossings.
【Over view】Lorem ipsum dolor sit amet, consectetur adipisicing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua. Ut enim ad minim veniam, quis nostrud exercitation ullamco laboris nisi ut aliquip ex ea commodo consequat. Duis aute irure dolor in reprehenderit in voluptate velit esse cillum dolore eu fugiat nulla pariatur. Excepteur sint occaecat cupidatat non proident, sunt in culpa qui officia deserunt mollit anim id est laborum.Lorem ipsum dolor sit amet, consec
[Usage with GaussView]
Able to use in the format of QST2/QST3 calculation of Gaussian.
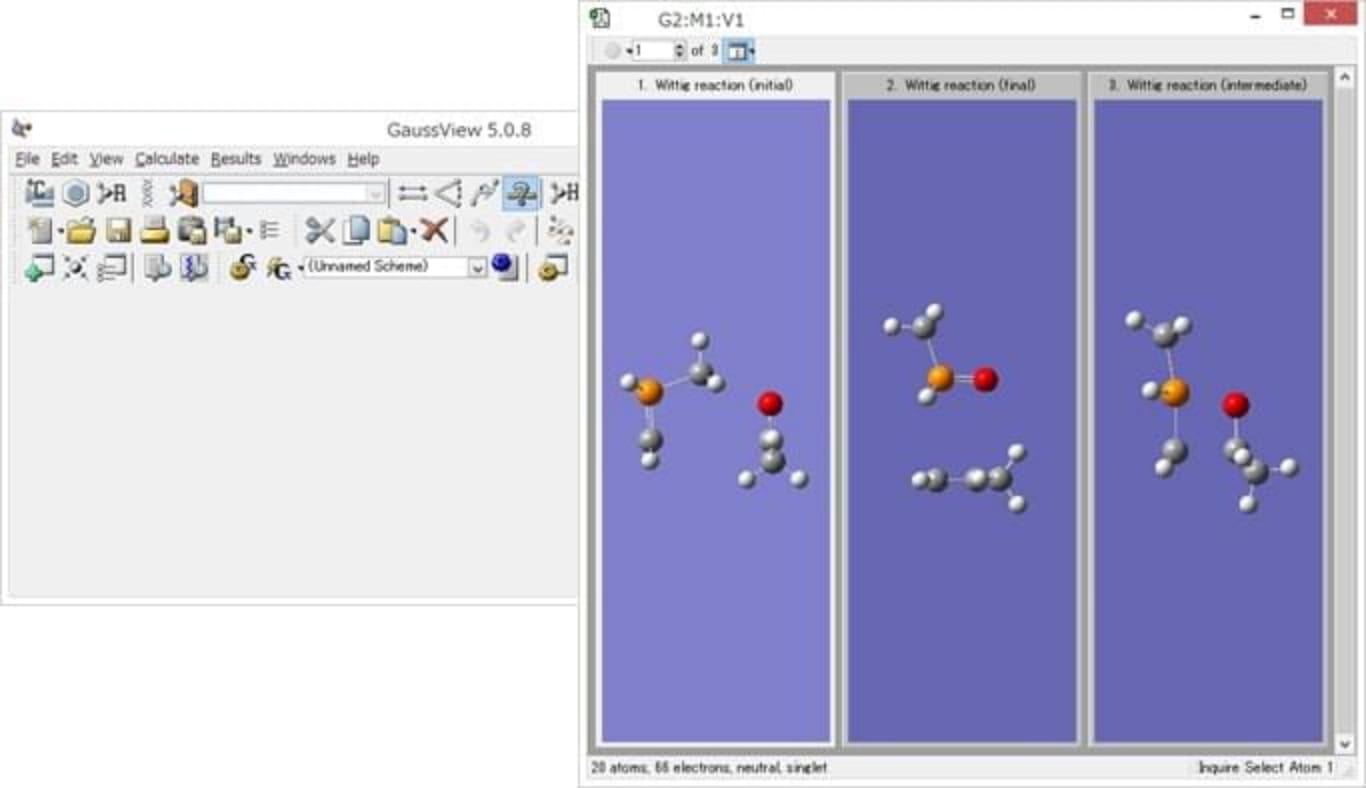
Input Creating with GaussView
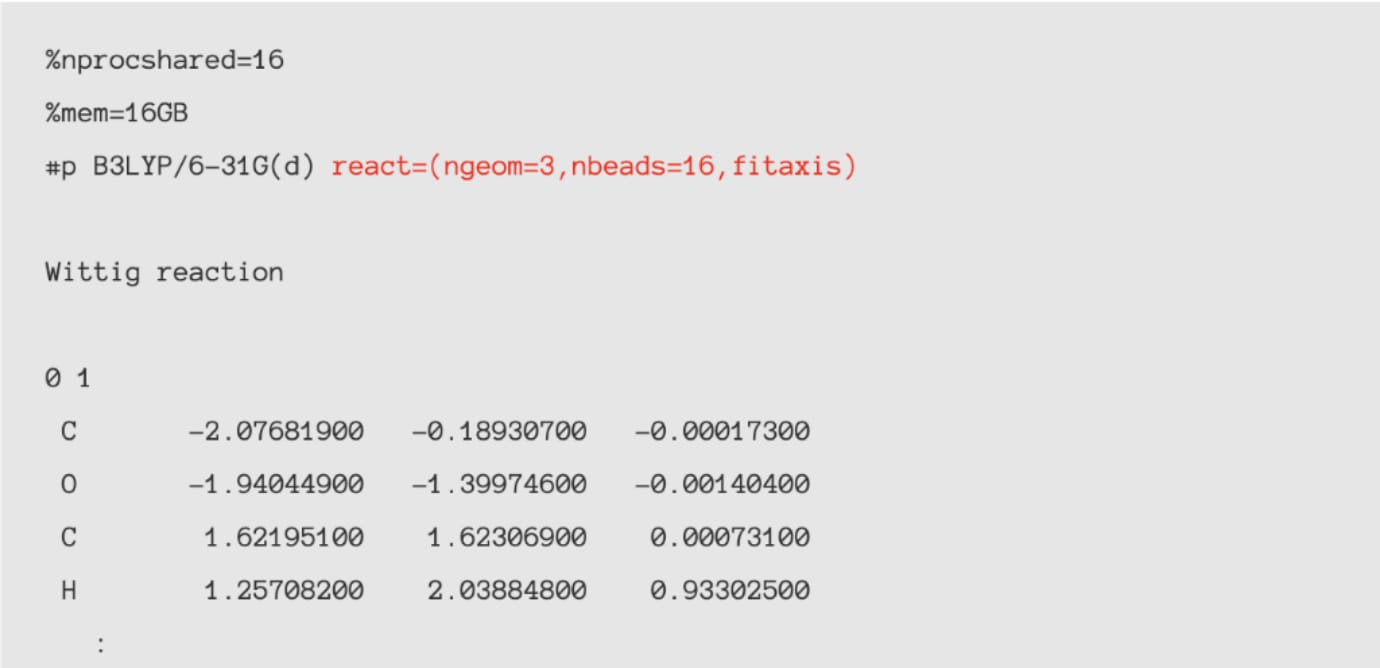
Contents of the input file to be created
The red part
is the original keyword for Reaction plus.
Three structures are specified: initial state,
intermediate state, final state
[Usage with AMBER / GROMACS]
ONIOM input-making support tool is attached to enables transforming
topology / coordinate file of AMBER and GROMACS to Gaussian format.

In addition, PDB and Mol2 file can be used as input file.
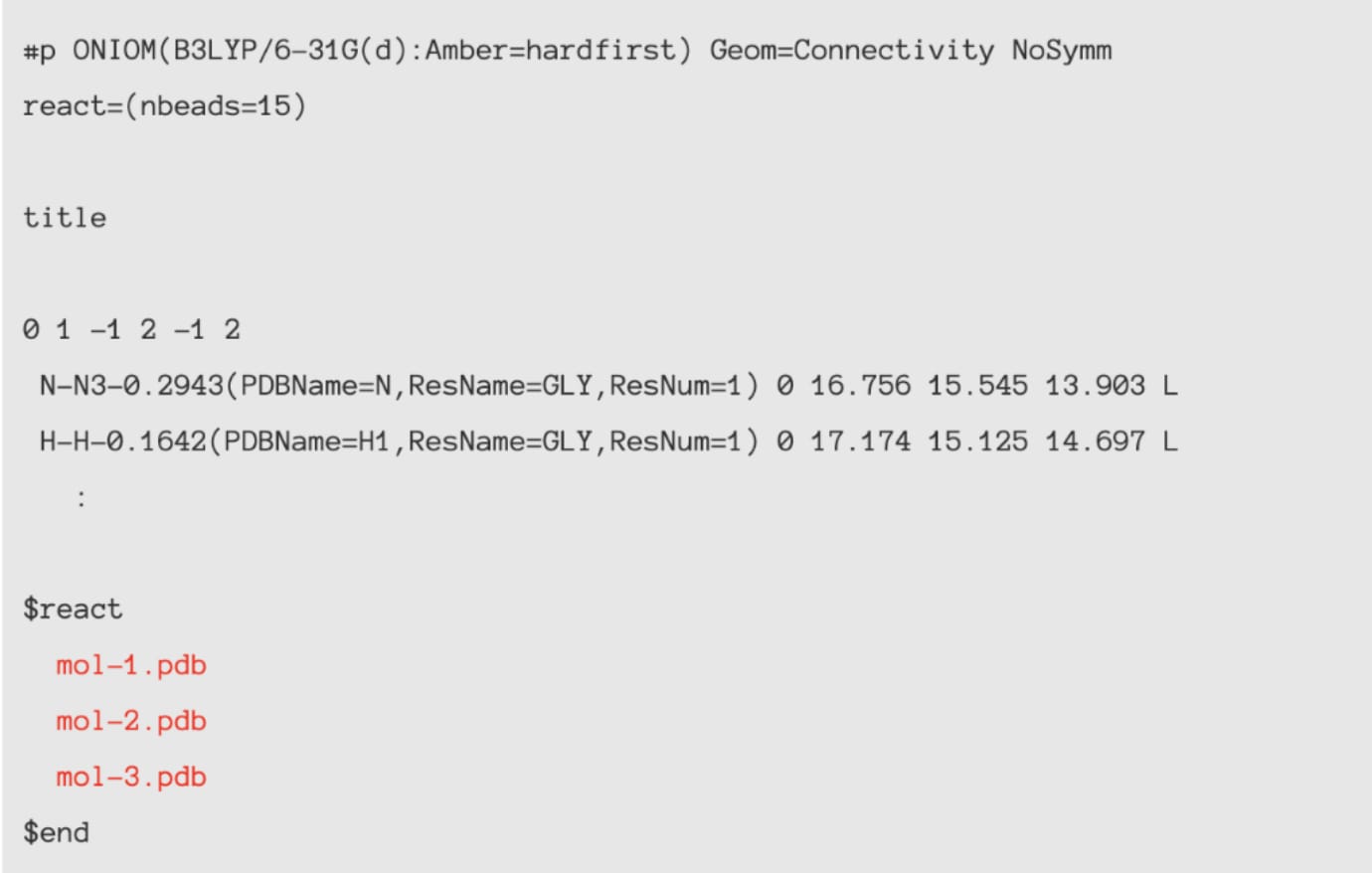
Specification of initial structure from PDB file
[Adapted to ONIOM method]
ONIOM method is suitable to ensure both calculation accuracy and
speed, which computes minutely to reaction active site but roughly to surroundings. By applying this
method, calculations of large-scale molecules such as enzyme reactions are now available.
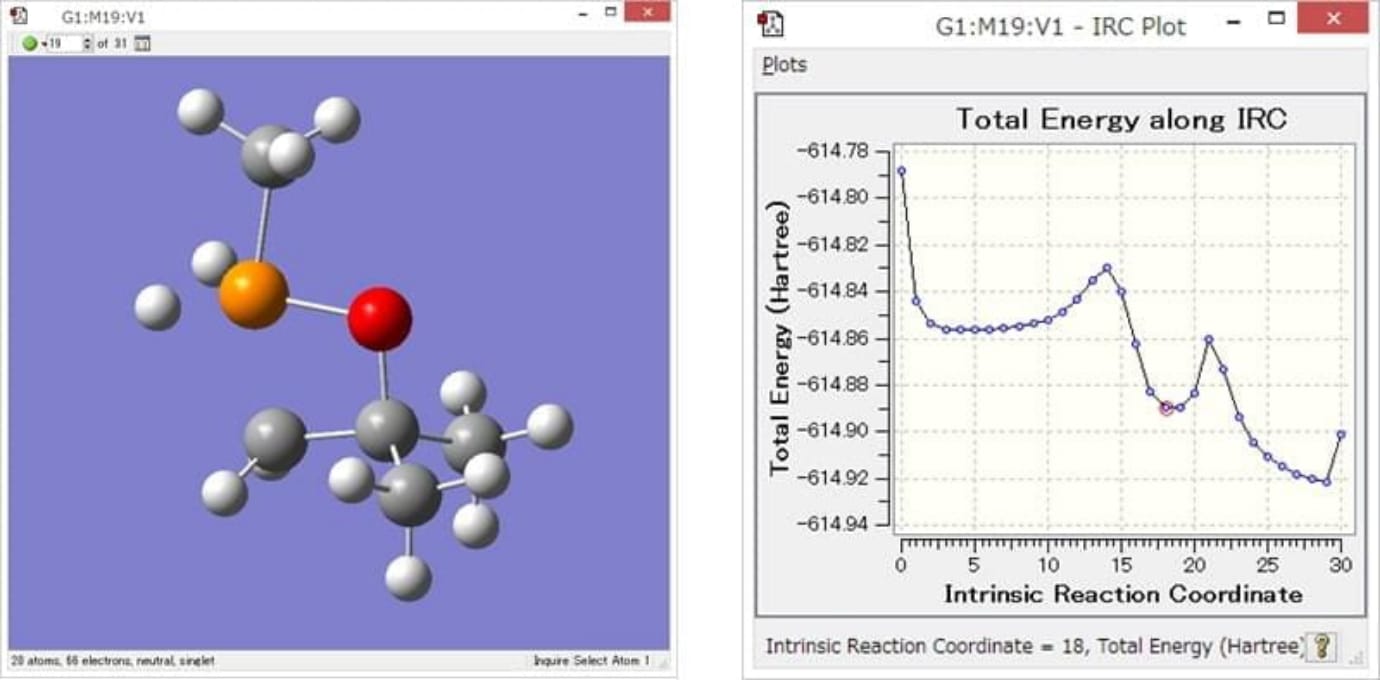
A Display example of GaussView
Chemical reaction animations and energy graphs can
be shown at a same time.
A tutorial is collectively attached, explaining how to create an input file in GaussView and how to
run the calculation (including Oniom calculation) with examples of organic chemical reactions as
well as frequently used techniques.
We also have a fee-based support service by E-mail in which
our staffs respond to various problems about techniques and solving the errors.
Case Studies
See example list
See publication list

*1 ONIOM Calculation Supporting Tool is used only for Linux.
*2 Parallelization
is not adapted between the nodes.
*3 This does not ensure the operation in all environment.
Especially, in the case of computing in Windows environments where security software are installed
causing the suspensions of calculations.

*1 Checked operation by GaussView 5.0.9 for Windows.
*2 ONIOM calculation
supporting tool can transform AMBER/GROMACS file into Gaussian (ONIOM) input (attached in Linux
version).
*3 log file can be read in animation with GaussView, while xyz and pdb file can be
displayed in animation with VMD or Avogadro (checked by GaussView 5.0.9, VMD 1.9.1 for Windows Avogadro
1.2.0 for Windows).
HPC SYSTEMS
Software license plans for academic or enterprise users are
respectively
available now
For more information, please contact us.
If there are any questions or troubles,
try to get in touch with us
through our
contact form (fee will be charged
depending on contents).
Released “Reaction plus Pro 2” , “Reaction plus Express 2”.
June 1st, 2018Released “Reaction plus Express”
May 1st, 2017Released original software “Reaction plus Pro”
January 1st, 2016